How repro-packs can save your future-self
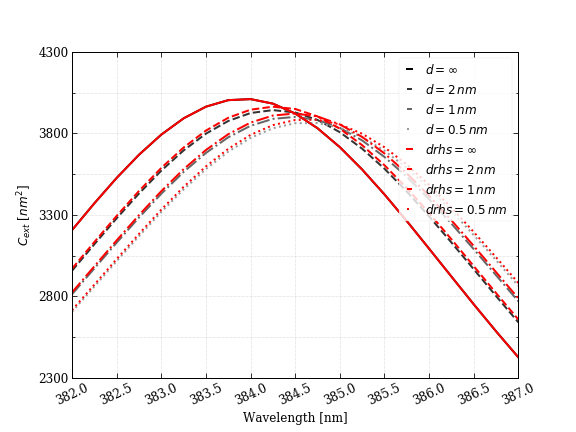
Extinction cross-section as a function of wavelength for an 8-nm silver sphere immersed in water with two BSA proteins placed at 2, 1, and 0.5 nm away from the surface in the z-direction, and at infinity (no protein). Red curves are after bug-fix.
The story of a bug-fix after the research paper was published.
Back in December 2019 we published a paper along with its reproducibility packages. These repro-packs, as we call them, consist of all the files necessary to reproduce the results in our paper (data and plots) and we deposit them in Zenodo and Figshare archives to obtain a citable DOI. The files are also kept in the paper repository, where we openly wrote the paper and worked on our replies to the reviewers. In the Barba Lab we believe this is the way of doing science and we work reproducibly staying true to the Reproducibility PI Manifesto
Here comes the twist… a couple of weeks ago (COVID-19 quarantine times) a student of our collaborator Christopher Cooper found a bug in PyGBe, our open source software used to obtain the results of the paper. He was in the process of implementing a new application, similar to the one in our paper, and he was following the math and code for inspiration. He was able to arrive at the same mathematical conclusions (phew...) but he was missing a term of the equations in the code. After reviewing the code over a video-call with our collaborator, we realized the student was right and that we had a bug!! We became very worried since this could mean having to contact the journal, and submitting a corrigendum with new results. We asked ourselves: how would this affect our conclusions in the paper?
First we had to fix the bug and then re-run all the simulations of the paper to see where we were standing to make a decision based on the results. But thanks Thor (or insert here God of preference) we had our repro-packs. After fixing the code, I went ahead and downloaded my own repro-packs (both execution and plotting files), and a few minutes later I was sending the simulations to run on a remote machine (remember we're in a pandemic). I had to send them in batches due to the hardware resources but after approximately a day and a half I had all the data I needed to compare my results.
How long do you think it would take you to get your simulations going to re-run a paper's results? In my case if I didn't have the repro-packs it would have take me from days to weeks. Once I had this new data, in just a few minutes I had all the new versions of the plots. Happy ending! The relevant quantity of the study (wavelength shift) remained intact, and even though the bug-fix affected the dependent variable (extinction cross section) by <1%, this had no impact on our conclusions.
Overall, the whole episode since the bug was fixed until I had our new results took between 2 and 3 days (including the time waiting for simulations to run). We confirmed that our conclusions didn't change and also added an errata to our paper repository in case someone wants to reproduce our results with the new code.
Working openly and in a reproducible way makes the whole research process more transparent. Making our research code open-source code allowed a person in another group to find our error, and making repro-packs saved my future-self (present today) from spending days or even weeks to get the results again after the bug-fix, and most of all I was able to have a quick diagnostic on the situation.
If this happens to you, how long do you think it can take you to reproduce all the figures in your paper?
Work reproducibly, it will save you and your supervisor headaches...
Natalia Clementi
PhD student in the Barba group