Lorena A. Barba group

Loading...
Accepted: Biomolecular electrostatics solver using Python and GPUs
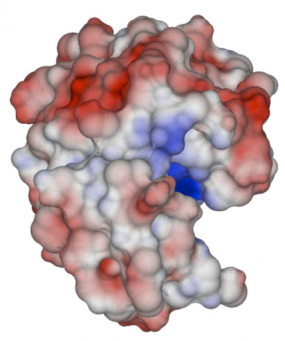
Submitted: 20 September, 2013. Accepted: 24 October, 2013. This paper presents a study of the effect of solvent-filled cavities and Stern layers in a biomolecular electrostatics solver based on a boundary integral formulation. The tool for this study was the PyGBe code: a solver for biomolecular electrostatics using Python, GPUs and boundary elements. To determine... Continue »
PyGBe: Python on the surface, GPUs at the heart. BEM solver for Electrostatics of Biomolecules
Poster presented at the GPU Technology Conference, March 2013, San Jose, CA.
It presents the PyGBe code (pronounced 'pig-bee'), which solves the linearized Poisson-Boltzmann equation using a boundary element method, BEM. The underlying dense systems are solved using a Krylov-subspace method, accelerated with... Continue »
Validation of the PyGBe code for Poisson-Boltzmann equation
The PyGBe code solves the linearized Poisson-Boltzmann equation using a boundary-integral formulation. We use a boundary element method with a collocation approach, and solve it via a Krylov-subspace method. To do this efficiently, the matrix-vector multiplications in the Krylov iterations are accelerated with a... Continue »