Probing protein orientation near charged surfaces
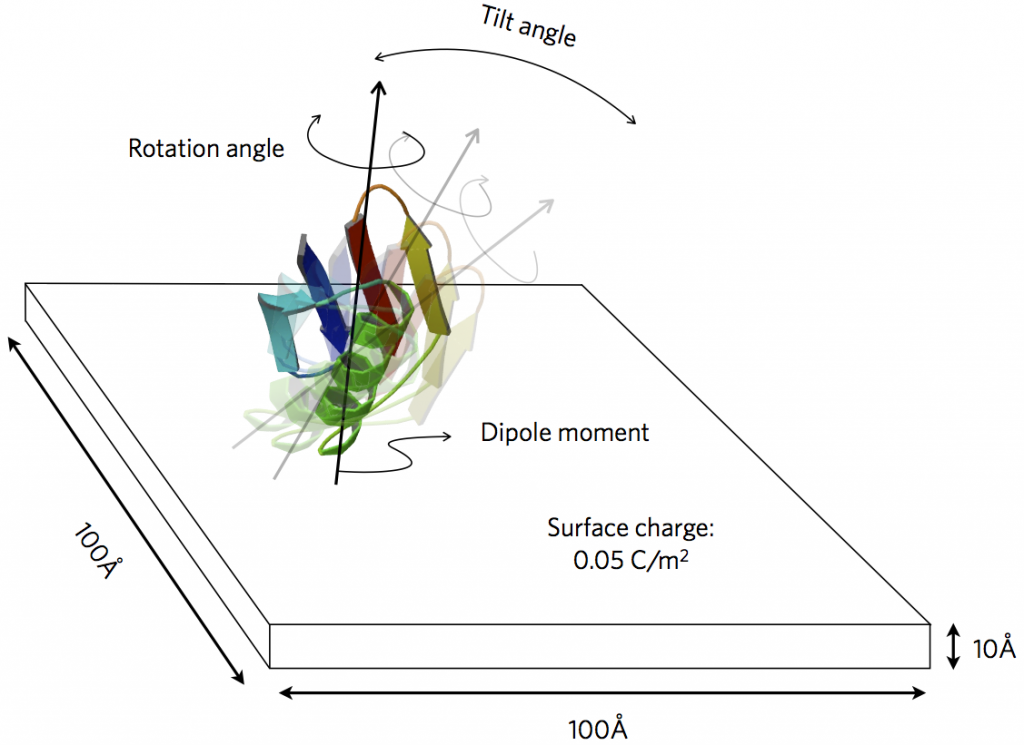
Figure from our paper on probing protein orientation near charged surfaces.
First version submitted: 31 March 2015. Submission of split papers: June 7 & June 12. Accepted: 4 Sept. & 26 Dec. 2015.
UPDATE: The material in the original manuscript was split in two papers, submitted to different peer-reviewed journals. The editorial decision on the first submission was that the paper was out of scope for the journal, which led us to reorganize the material into a paper on the code extension and grid-convergence study (for a computational journal) and the protein adsorption studies (for a chemical physics journal).
After submission for peer review, and with the pre-prints on arXiv, we also made public the manuscript source files in the GitHub repository
Title 1
- "Probing protein orientation near charged nanosurfaces for simulation-assisted biosensor design", Christopher D. Cooper, Natalia C. Clementi, Lorena A. Barba. J. Chem. Phys., 143:124709 (September 2015). 10.1063/1.4931113 // Preprint arXiv:1503.08150v4 // code repository // manuscript repository // figshare: orientation of protein GB1 // figshare: orientation of immunoglobulin G // figshare: grid convergence, immunoglobulin G
Version 3 of the preprint is the version submitted to a journal, after the original manuscript was split in two. Version 4 is the post peer-review revision.
Title 2
- "Poisson-Boltzmann model for protein-surface electrostatic interactions and grid-convergence study using the PyGBe code", Christopher D. Cooper, Lorena A. Barba. Comput. Phys. Comm., 202:23-32 (May 2016). 10.1016/j.cpc.2015.12.019 // Preprint arXiv:1506.03745 // Code repository // manuscript repository // figshare: grid convergence, spherical surfaces // figshare: grid convergence on protein GB1
Accepted 26 Dec. 2015
Overview
These new papers present our latest research using PyGBe, the open-source code for Poisson-Boltzmann equations using the boundary element method, algorithmic acceleration via multipole expansions and hardware acceleration via GPUs.
We have extended PyGBe to treat the interaction of molecules with engineered surfaces of prescribed charge or electric potential. The application of interest is biosensors, where performance is significantly affected by the orientation of ligand molecules near the bioactive surface. A sensor element (functionalized with a self-assembled monolayer) can be represented as a charged surface, interacting with a biomolecule through electrostatics. We can thus study the preferred orientation of certain proteins under various conditions of prescribed surface charge and ambient salt concentration.
To be able to verify code correctness, we developed an analytical solution for two spheres interacting electrostatically, where one of them has an imposed charge. We used it to complete a grid convergence analysis. Then, we completed two studies of protein adsorption: one with protein GB1 D4' (mutant), and the other with immunoglobulin G. In the first case, we were able to compare with published results, both experimental and with molecular dynamics. In the second—of greater relevance for biosensors—we studied the preferred orientation of immunoglobulin G under various ambient conditions. Through this study, we concluded that this protein is best immobilized on a surface with high (positive) surface charge and low salt concentration.
Open data
- "Protein orientation near charged surface using PyGBe with immunoglobulin G", Christopher D. Cooper, Lorena A. Barba. (March 2015). 10.6084/m9.figshare.1348802.v8 // Paper containing figure(s)
Reproducibility package including data, running script, plotting script and figure. On figshare under CC-BY.
- "Grid convergence of PyGBe with immunoglobulin G near a charged surface", Christopher D. Cooper, Lorena A. Barba. (March 2015). 10.6084/m9.figshare.1348801.v8 // Paper containing figure
Reproducibility package including data, running script, plotting script and figure. On figshare under CC-BY.
- "Protein orientation near a charged surface using PyGBe and protein G B1 D4'", Christopher D. Cooper, Lorena A. Barba. (March 2015). https://dx.doi.org/10.6084/m9.figshare.1348804.v6 // Paper containing figure(s)
Reproducibility package including data, running script, plotting script and figure. On figshare under CC-BY.
- "Grid convergence of PyGBe with protein G B1 D4'", Christopher D. Cooper, Lorena A. Barba. (March 2015). 10.6084/m9.figshare.1348803.v10 // Paper containing figure
Reproducibility package including data, running script, plotting script and figure(s). On figshare under CC-BY.
- "Grid convergence of PyGBe with a spherical molecule near spherical surface", Christopher D. Cooper, Lorena A. Barba. (March 2015). 10.6084/m9.figshare.1348841.v4 // Paper containing figure
Reproducibility package including data, running script, plotting script, and figure of grid-convergence study using two spheres, one with prescribed potential. On figshare under CC-BY.
Acknowledgements
This work was supported by ONR via grant #N00014-11-1-0356 of the Applied Computational Analysis Program. LAB also acknowledges support from NSF CAREER award OCI-1149784 and from NVIDIA, Inc. via the CUDA Fellows Program.
We are grateful for many helpful conversations with members of the Materials and Sensors Branch of the Naval Research Laboratory, especially Dr. Jeff M. Byers and Dr. Marc Raphael.